R6 Class for Gage R&R (Repeatability and Reproducibility) Analysis
Public fields
XData frame containing the measurement data.
ANOVAList containing the results of the Analysis of Variance (ANOVA) for the gage study.
RedANOVAList containing the results of the reduced ANOVA.
methodCharacter string specifying the method used for the analysis (e.g.,
`crossed`,`nested`).EstimatesList of estimates including variance components, repeatability, and reproducibility.
VarcompList of variance components.
SigmaNumeric value representing the standard deviation of the measurement system.
GageNameCharacter string representing the name of the gage.
GageToleranceNumeric value indicating the tolerance of the gage.
DateOfStudyCharacter string representing the date of the gage R&R study.
PersonResponsibleCharacter string indicating the person responsible for the study.
CommentsCharacter string for additional comments or notes about the study.
bFactor levels for operator.
aFactor levels for part.
yNumeric vector or matrix containing the measurement responses.
facNamesCharacter vector specifying the names of the factors (e.g.,
`Operator`,`Part`).numOInteger representing the number of operators.
numPInteger representing the number of parts.
numMInteger representing the number of measurements per part-operator combination.
Methods
Method new()
Initialize the fiels of the gageRR object
Usage
gageRR.c$new(
X,
ANOVA = NULL,
RedANOVA = NULL,
method = NULL,
Estimates = NULL,
Varcomp = NULL,
Sigma = NULL,
GageName = NULL,
GageTolerance = NULL,
DateOfStudy = NULL,
PersonResponsible = NULL,
Comments = NULL,
b = NULL,
a = NULL,
y = NULL,
facNames = NULL,
numO = NULL,
numP = NULL,
numM = NULL
)Arguments
XData frame containing the measurement data.
ANOVAList containing the results of the Analysis of Variance (ANOVA) for the gage study.
RedANOVAList containing the results of the reduced ANOVA.
methodCharacter string specifying the method used for the analysis (e.g., "crossed", "nested").
EstimatesList of estimates including variance components, repeatability, and reproducibility.
VarcompList of variance components.
SigmaNumeric value representing the standard deviation of the measurement system.
GageNameCharacter string representing the name of the gage.
GageToleranceNumeric value indicating the tolerance of the gage.
DateOfStudyCharacter string representing the date of the gage R&R study.
PersonResponsibleCharacter string indicating the person responsible for the study.
CommentsCharacter string for additional comments or notes about the study.
bFactor levels for operator.
aFactor levels for part.
yNumeric vector or matrix containing the measurement responses.
facNamesCharacter vector specifying the names of the factors (e.g., "Operator", "Part").
numOInteger representing the number of operators.
numPInteger representing the number of parts.
numMInteger representing the number of measurements per part-operator combination.
Method subset()
Return a subset of the data frame that containing the measurement data (X)
Method summary()
Summarize the information of the fields of the gageRR object.
Method as.data.frame()
Methods for function as.data.frame in Package base.
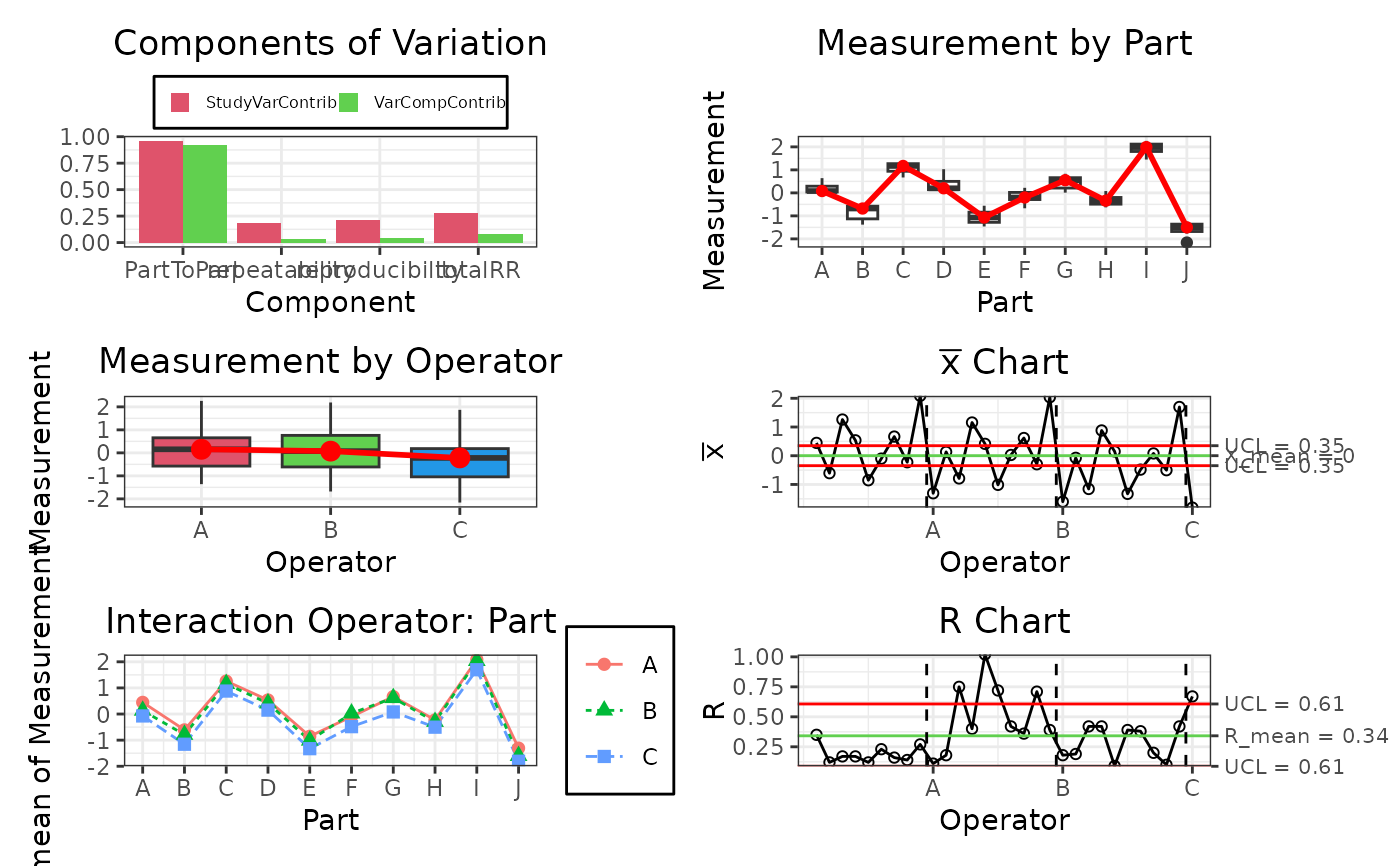
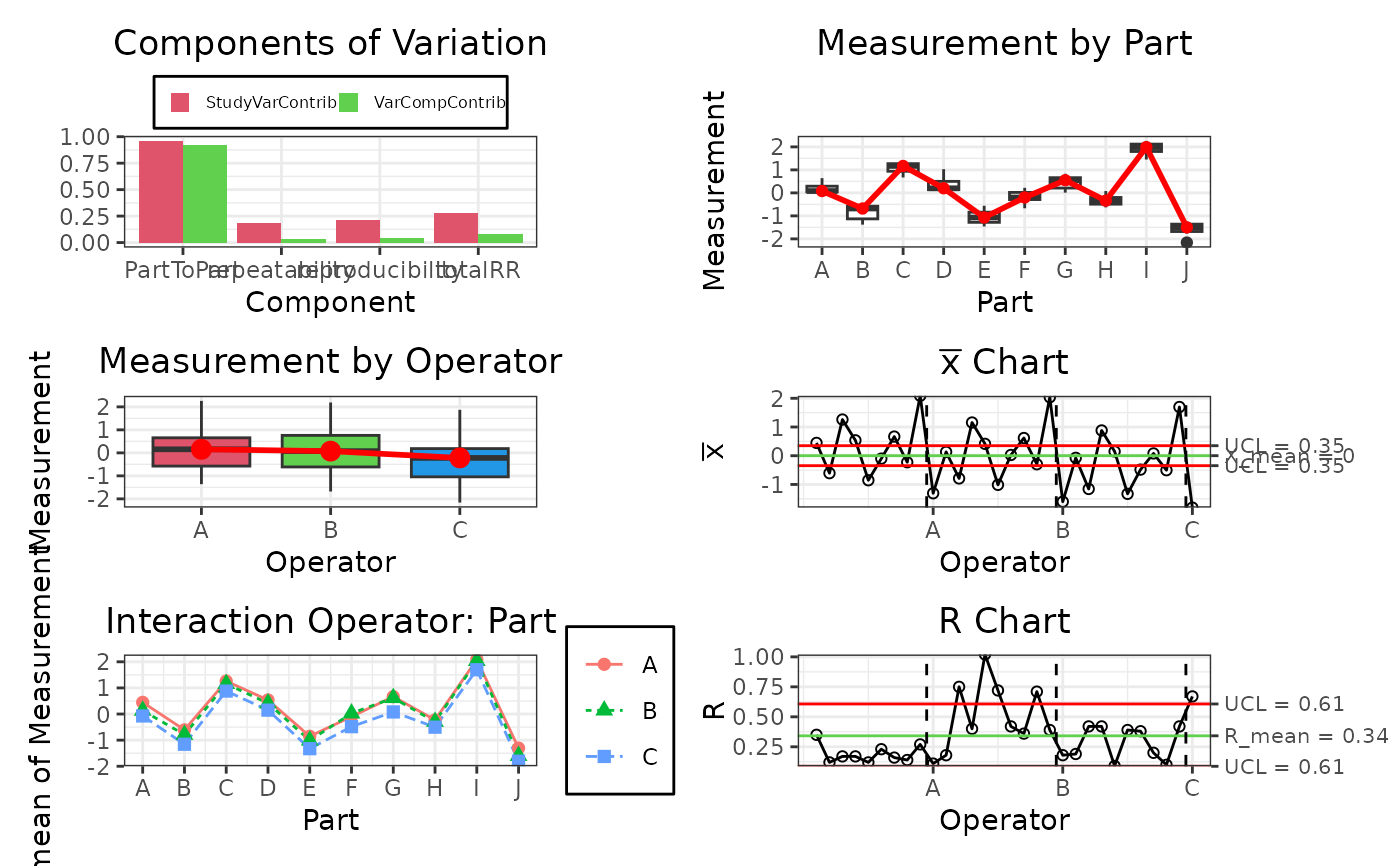
Method plot()
This function creates a customized plot using the data from the gageRR.c object.
Arguments
mainCharacter string specifying the title of the plot.
xlabA character string for the x-axis label.
ylabA character string for the y-axis label.
colA character string or vector specifying the color(s) to be used for the plot elements.
lwdA numeric value specifying the line width of plot elements
funFunction to use for the calculation of the interactions (e.g.,
mean,median). Default ismean.
Examples
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
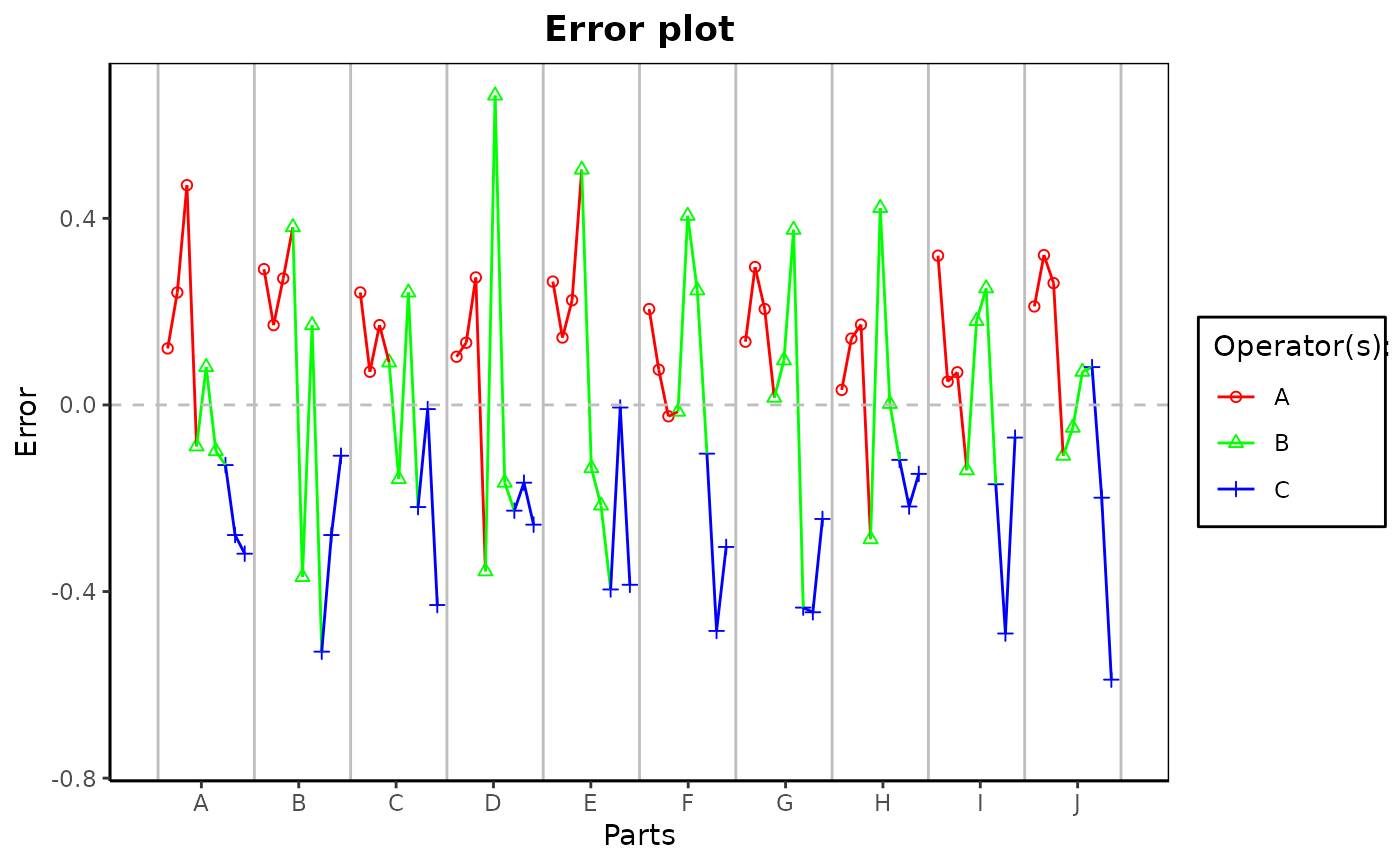
gdo$plot()Method errorPlot()
The data from an object of class gageRR can be analyzed by running `Error Charts` of the individual deviations from the accepted rference values. These `Error Charts` are provided by the function errorPlot.
Arguments
maina main title for the plot.
xlabA character string for the x-axis label.
ylabA character string for the y-axis label.
colPlotting color.
pchAn integer specifying a symbol or a single character to be used as the default in plotting points.
ylimThe y limits of the plot.
legendA logical value specifying whether a legend is plotted automatically. By default legend is set to `TRUE`.
Examples
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
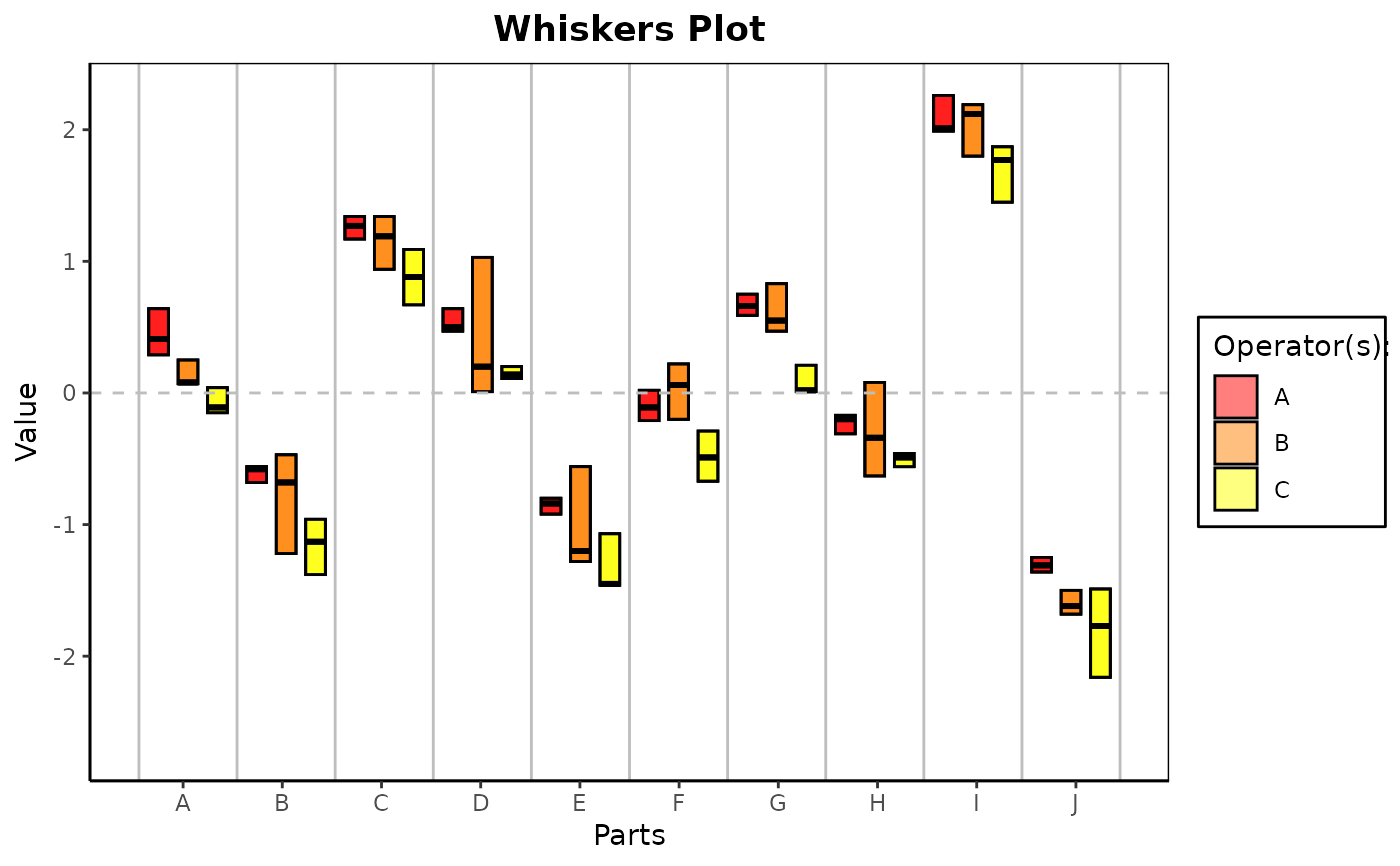
gdo$errorPlot()Method whiskersPlot()
In a Whiskers Chart, the high and low data values and the average (median) by part-by-operator are plotted to provide insight into the consistency between operators, to indicate outliers and to discover part-operator interactions. The Whiskers Chart reminds of boxplots for every part and every operator.
Arguments
maina main title for the plot.
xlabA character string for the x-axis label.
ylabA character string for the y-axis label.
colPlotting color.
ylimThe y limits of the plot.
legendA logical value specifying whether a legend is plotted automatically. By default legend is set to `TRUE`.
Examples
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
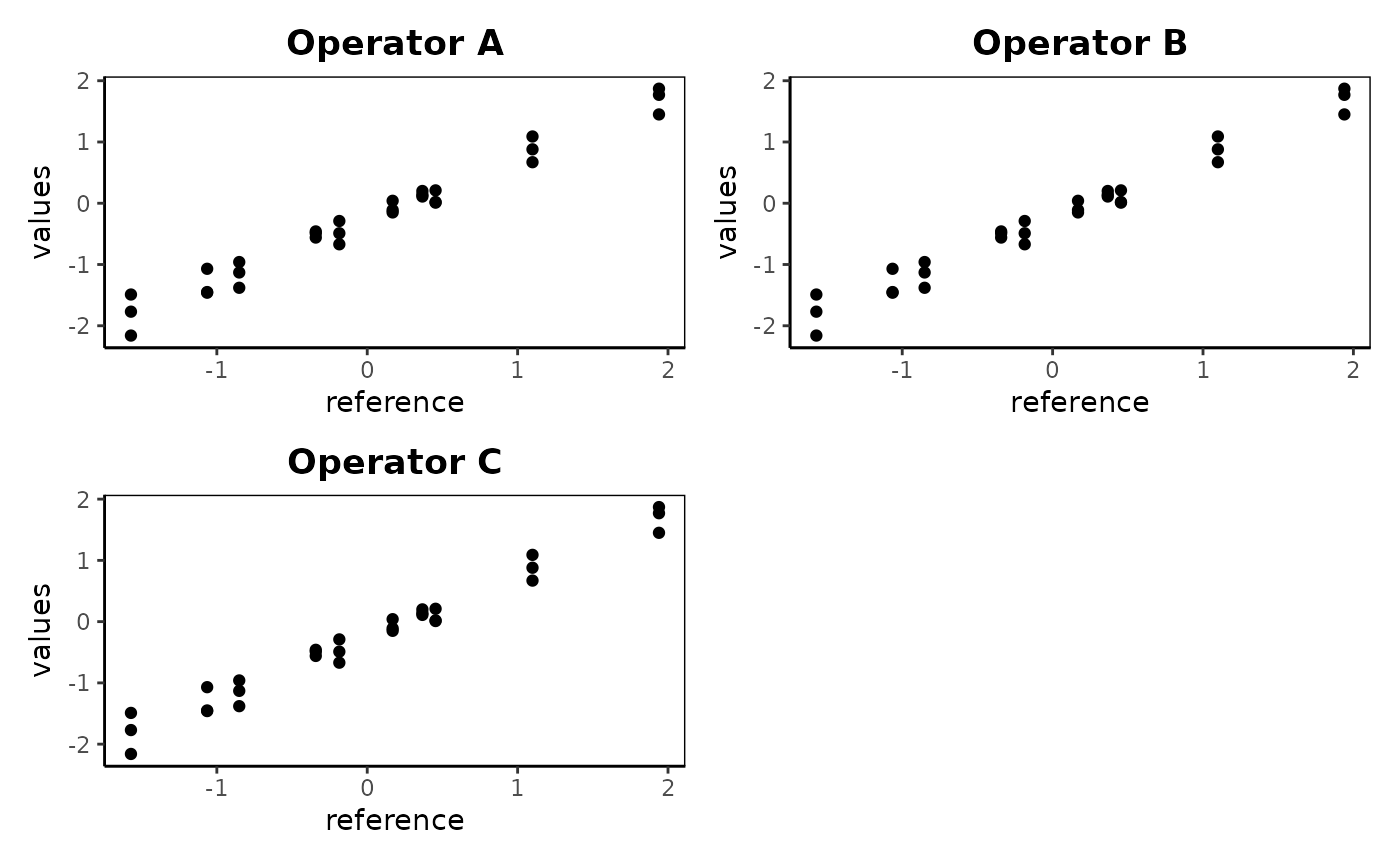
gdo$whiskersPlot()Method averagePlot()
averagePlot creates all x-y plots of averages by size out of an object of class gageRR. Therfore the averages of the multiple readings by each operator on each part are plotted with the reference value or overall part averages as the index.
Arguments
maina main title for the plot.
xlabA character string for the x-axis label.
ylabA character string for the y-axis label.
colPlotting color.
singleA logical value.If `TRUE` a new graphic device will be opened for each plot. By default
singleis set to `FALSE`.
Examples
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
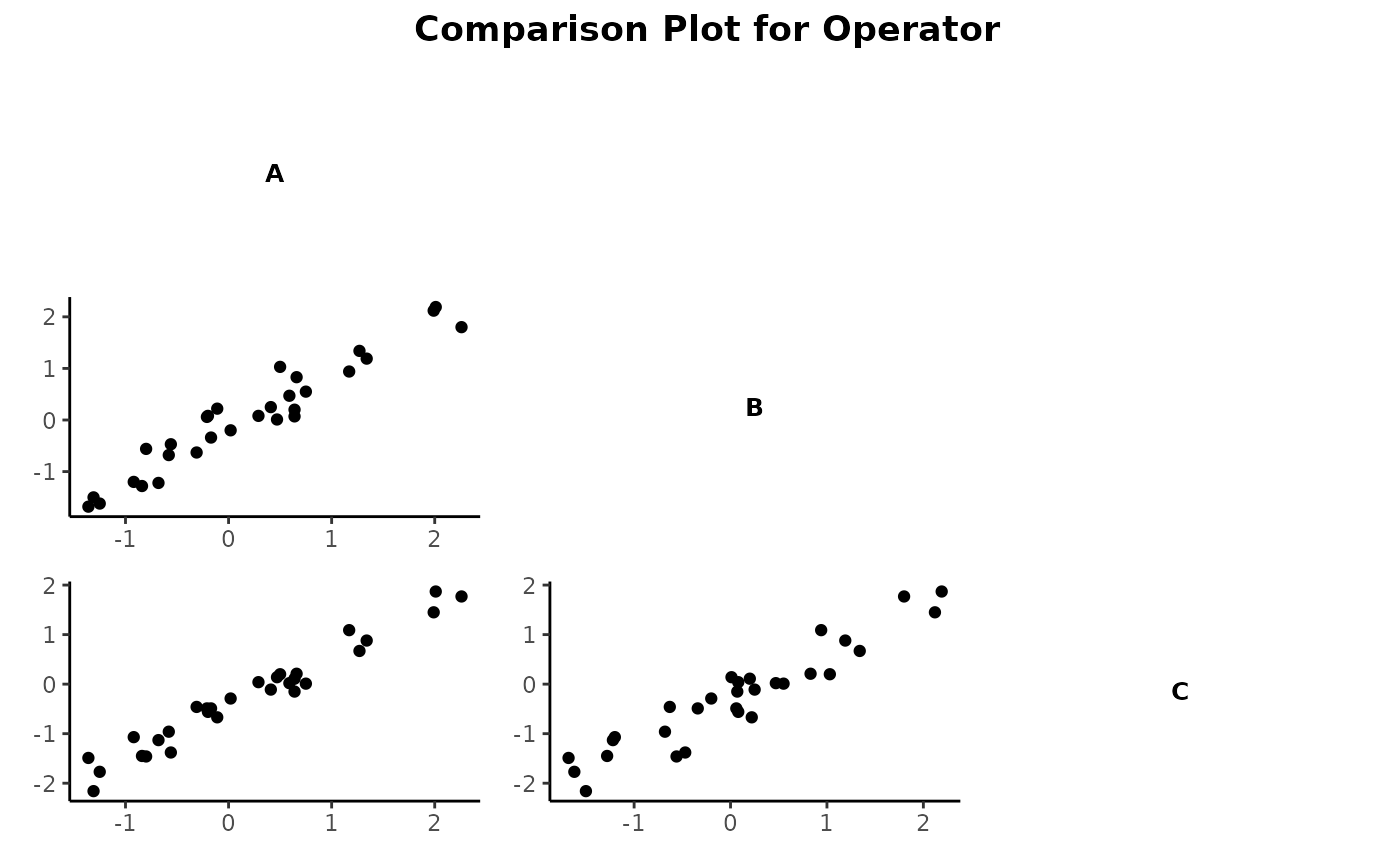
gdo$averagePlot()Method compPlot()
compPlot creates comparison x-y plots of an object of class gageRR. The averages of the multiple readings by each operator on each part are plotted against each other with the operators as indices. This plot compares the values obtained by one operator to those of another.
Arguments
maina main title for the plot.
xlabA character string for the x-axis label.
ylabA character string for the y-axis label.
colPlotting color.
cex.labThe magnification to be used for x and y labels relative to the current setting of cex.
funOptional function that will be applied to the multiple readings of each part. fun should be an object of class
functionlikemean,median,sum, etc. By default,funis set to `NULL` and all readings will be plotted.
Examples
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
gdo$compPlot()Examples
#create gageRR-object
gdo <- gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
#vector of responses
y <- c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
#appropriate responses
gdo$response(y)
# perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
# Using the plots
gdo$plot()
 ## ------------------------------------------------
## Method `gageRR.c$plot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$plot()
## ------------------------------------------------
## Method `gageRR.c$plot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$plot()
 ## ------------------------------------------------
## Method `gageRR.c$errorPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$errorPlot()
## ------------------------------------------------
## Method `gageRR.c$errorPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$errorPlot()
 ## ------------------------------------------------
## Method `gageRR.c$whiskersPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$whiskersPlot()
## ------------------------------------------------
## Method `gageRR.c$whiskersPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$whiskersPlot()
 ## ------------------------------------------------
## Method `gageRR.c$averagePlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$averagePlot()
## ------------------------------------------------
## Method `gageRR.c$averagePlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$averagePlot()
 ## ------------------------------------------------
## Method `gageRR.c$compPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$compPlot()
## ------------------------------------------------
## Method `gageRR.c$compPlot`
## ------------------------------------------------
# Create gageRR-object
gdo = gageRRDesign(Operators = 3, Parts = 10, Measurements = 3, randomize = FALSE)
# Vector of responses
y = c(0.29,0.08, 0.04,-0.56,-0.47,-1.38,1.34,1.19,0.88,0.47,0.01,0.14,-0.80,
-0.56,-1.46, 0.02,-0.20,-0.29,0.59,0.47,0.02,-0.31,-0.63,-0.46,2.26,
1.80,1.77,-1.36,-1.68,-1.49,0.41,0.25,-0.11,-0.68,-1.22,-1.13,1.17,0.94,
1.09,0.50,1.03,0.20,-0.92,-1.20,-1.07,-0.11, 0.22,-0.67,0.75,0.55,0.01,
-0.20, 0.08,-0.56,1.99,2.12,1.45,-1.25,-1.62,-1.77,0.64,0.07,-0.15,-0.58,
-0.68,-0.96,1.27,1.34,0.67,0.64,0.20,0.11,-0.84,-1.28,-1.45,-0.21,0.06,
-0.49,0.66,0.83,0.21,-0.17,-0.34,-0.49,2.01,2.19,1.87,-1.31,-1.50,-2.16)
# Appropriate responses
gdo$response(y)
# Perform and gageRR
gdo <- gageRR(gdo)
#>
#> AnOVa Table - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 34.440 1.09e-10 ***
#> Part 9 88.36 9.818 213.517 < 2e-16 ***
#> Operator:Part 18 0.36 0.020 0.434 0.974
#> Residuals 60 2.76 0.046
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#> AnOVa Table Without Interaction - crossed Design
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Operator 2 3.17 1.584 39.62 1.34e-12 ***
#> Part 9 88.36 9.818 245.61 < 2e-16 ***
#> Residuals 78 3.12 0.040
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> ----------
#>
#> Gage R&R
#> VarComp VarCompContrib Stdev StudyVar StudyVarContrib
#> totalRR 0.0914 0.0776 0.302 1.81 0.279
#> repeatability 0.0400 0.0339 0.200 1.20 0.184
#> reproducibility 0.0515 0.0437 0.227 1.36 0.209
#> Operator 0.0515 0.0437 0.227 1.36 0.209
#> Operator:Part 0.0000 0.0000 0.000 0.00 0.000
#> Part to Part 1.0864 0.9224 1.042 6.25 0.960
#> totalVar 1.1779 1.0000 1.085 6.51 1.000
#>
#> ---
#> * Contrib equals Contribution in %
#> **Number of Distinct Categories (truncated signal-to-noise-ratio) = 4
#>
gdo$compPlot()